
Steven Lee
Cellular players get their moment in the limelight
In order to understand our biology, researchers need to investigate not only what cells are doing, but also more specifically what is happening inside of cells at the level of organelles, the specialized structures that perform unique tasks to keep the cell functioning. However, most methods for analysis take place at the level of the whole cell. Because a specific organelle might make up only a fraction of an already microscopic cell’s contents, “background noise” from other cellular components can drown out useful information about the organelle being studied, such as changes in the organelle’s protein or metabolite levels in response to different conditions.
Whitehead Institute Member David Sabatini and Walter Chen, a former graduate student in Sabatini’s lab and now a pediatrics resident at Boston Children’s Hospital and Boston Medical Center and a postdoctoral researcher at Harvard Medical School, developed in recent years a method for isolating organelles for analysis that outstrips previous methods in its ability to purify organelles both rapidly and specifically. They first applied the method to mitochondria, the energy-generating organelles known as the “powerhouses of the cell,” and published their study in Cell in 2016. Subsequently, former Sabatini lab postdoctoral researcher Monther Abu-Remaileh and graduate student Gregory Wyant applied the method to lysosomes, the recycling plants of cells that break down cell parts for reuse, as described in the journal Science in 2017. In collaboration with former Sabatini lab postdoctoral researcher Kivanc Birsoy, Sabatini and Chen next developed a way to use the mitochondrial method in mice, as described in PNAS in 2019. Now, in a paper published in iScience on May 22, Sabatini, Chen, and graduate student Jordan Ray have extended the method for use on peroxisomes, organelles that play essential roles in human physiology.
“It’s gratifying to see this toolkit expand so we can use it to gain insight into the nuances of these organelles’ biology,” Sabatini says.
Using their organellar immunoprecipitation techniques, the researchers have uncovered previously unknown aspects of mitochondrial biology, including changes in metabolites during diverse states of mitochondrial function. They also uncovered new aspects of lysosomal biology, including how nutrient starvation affects the exchange of amino acids between the organelle and the rest of the cell. Their methods could help researchers gain new insights into diseases in which mitochondria or lysosomes are affected, such as mitochondrial respiratory chain disorders, lysosomal storage diseases, and Parkinson’s Disease. Now that Sabatini, Chen, and Ray have extended the method to peroxisomes, it could also be used to learn more about peroxisome-linked disorders.
Developing a potent method
The researchers’ method is based on “organellar immunoprecipitation,” which utilizes antibodies, immune system proteins that recognize specific perceived threats that they are supposed to bind to and help remove from the body. The researchers create a custom tag for each type of organelle by taking an epitope, the section of a typical perceived threat that antibodies recognize and bind to, and fusing it to a protein that is known to localize to the membrane of the organelle of interest, so the tag will attach to the organelle. The cells containing these tagged organelles are first broken up to release all of the cell’s contents, and then put in solution with tiny magnetic beads covered in the aforementioned antibodies. The antibodies on the beads latch onto the tagged organelles. A magnet is then used to collect all of the beads and separate the bound organelles from the rest of the cellular material, while contaminants are washed away. The resulting isolated organelles can subsequently be analyzed using a variety of methods that look at the organelles’ metabolites, lipids, and proteins.
With their method, Chen and Sabatini have developed an organellar isolation technique that is both rapid and specific, qualities that prior methods have typically lacked. The workflow that Chen and Sabatini developed is fast—this new iteration for peroxisomes takes only 10 minutes to isolate the tagged organelles once they have been released from cells. Speed is important because the natural profile of the organelles’ metabolites and proteins begins to change once they are released from the cell, and the longer the process takes, the less the results will reflect the organelle’s native state.
“We’re interested in studying the metabolic contents of organelles, which can be labile over the course of an isolation,” Chen says. “Because of their speed and specificity, these methods allow us to not only better assess the original metabolic profile of a specific organelle but also study proteins that may have more transient interactions with the organelle, which is very exciting.”
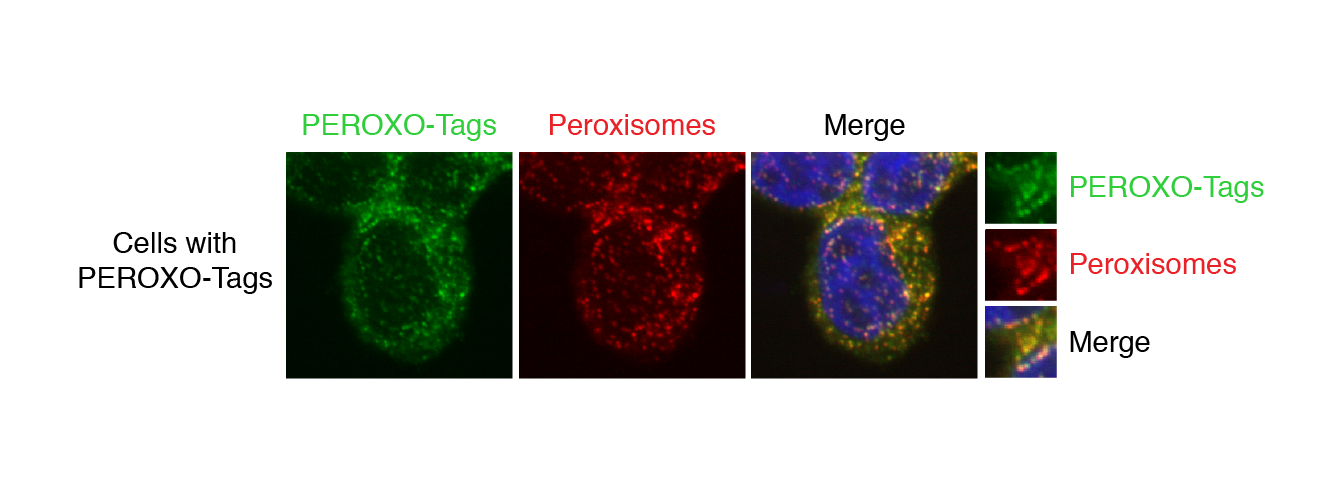
Peroxisomes take the limelight
Peroxisomes are organelles that are important for multiple metabolic processes and contribute to a number of essential biological functions, such as producing the insulating myelin sheaths for neurons. Defects in peroxisomal function are found in various genetic disorders in children and have been implicated in neurodegenerative diseases as well. However, compared to other organelles such as mitochondria, peroxisomes are relatively understudied. Being able to get a close-up look at the contents of peroxisomes may provide insights into important and previously unappreciated biology. Importantly, in contrast to traditional ways of isolating peroxisomes, the new method that Sabatini, Chen, and Ray have developed is not only fast and specific, but also reproducible and easy to use.
“Peroxisomal biology is quite fascinating, and there are a lot of outstanding questions about how they are formed, how they mature, and what their role is in disease that hopefully this tool can help elucidate,” Ray says.
An exciting next step may be to adapt the peroxisome isolation method so it can be used in a mammaliam model organism, such as mice, something the researchers have already done with the mitochondrial version.
“Using this method in animals could be especially helpful for studying peroxisomes because peroxisomes participate in a variety of functions that are essential on an organismal rather than cellular level,” Chen says. Going forward, Chen is interested in using the method to profile the contents of peroxisomes in specific cell types across a panel of different mammalian organs.
While Chen sets out to discover what unknown biology the peroxisome isolation method can reveal, researchers in Sabatini’s lab are busy working on another project: extending the method to even more organelles.
***
David Sabatini's primary affiliation is with Whitehead Institute for Biomedical Research, where his laboratory is located and all his research is conducted. He is also a Howard Hughes Medical Institute investigator and a professor of biology at Massachusetts Institute of Technology.
***
Citations:
G. Jordan Ray, Elizabeth A. Boydston, Emily Shortt, Gregory A. Wyant, Sebastian Lourido, Walter W. Chen, David M. Sabatini, “A PEROXO-Tag Enables Rapid Isolation of Peroxisomes from Human Cells,” iScience, May 22, 2020.
Bayraktar et al., “MITO-Tag Mice enable rapid isolation and multimodal profiling of mitochondria from specific cell types in vivo,” PNAS, Jan 2, 2019.
Abu-Remaileh et al., “Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes,” Science, Nov 10, 2017.
Chen et al., “Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism,” Cell, August 25, 2016.
Topics
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


